Seacows Growth, Bayesian Example
Data and Model
seacows=list(x=c(1.0,1.5,1.5,1.5,2.5,4.0,5.0,5.0,7.0,8.0,
8.5,9.0,9.5,9.5,10.0,12.0,12.0,13.0,13.0,14.5,
15.5,15.5,16.5,17.0,22.5,29.0,31.5),y=c(1.80,
1.85,1.87,1.77,2.02,2.27,2.15,2.26,2.47,2.19,
2.26,2.40,2.39,2.41,2.50,2.32,2.32,2.43,2.47,
2.56,2.65,2.47,2.64,2.56,2.70,2.72,2.57),N=27)
library(plyr)
dt=t(ldply(list(seacows[[1]],seacows[[2]])))
colnames(dt)=c("x","y")
dt=as.data.frame(dt)
library(ggplot2)
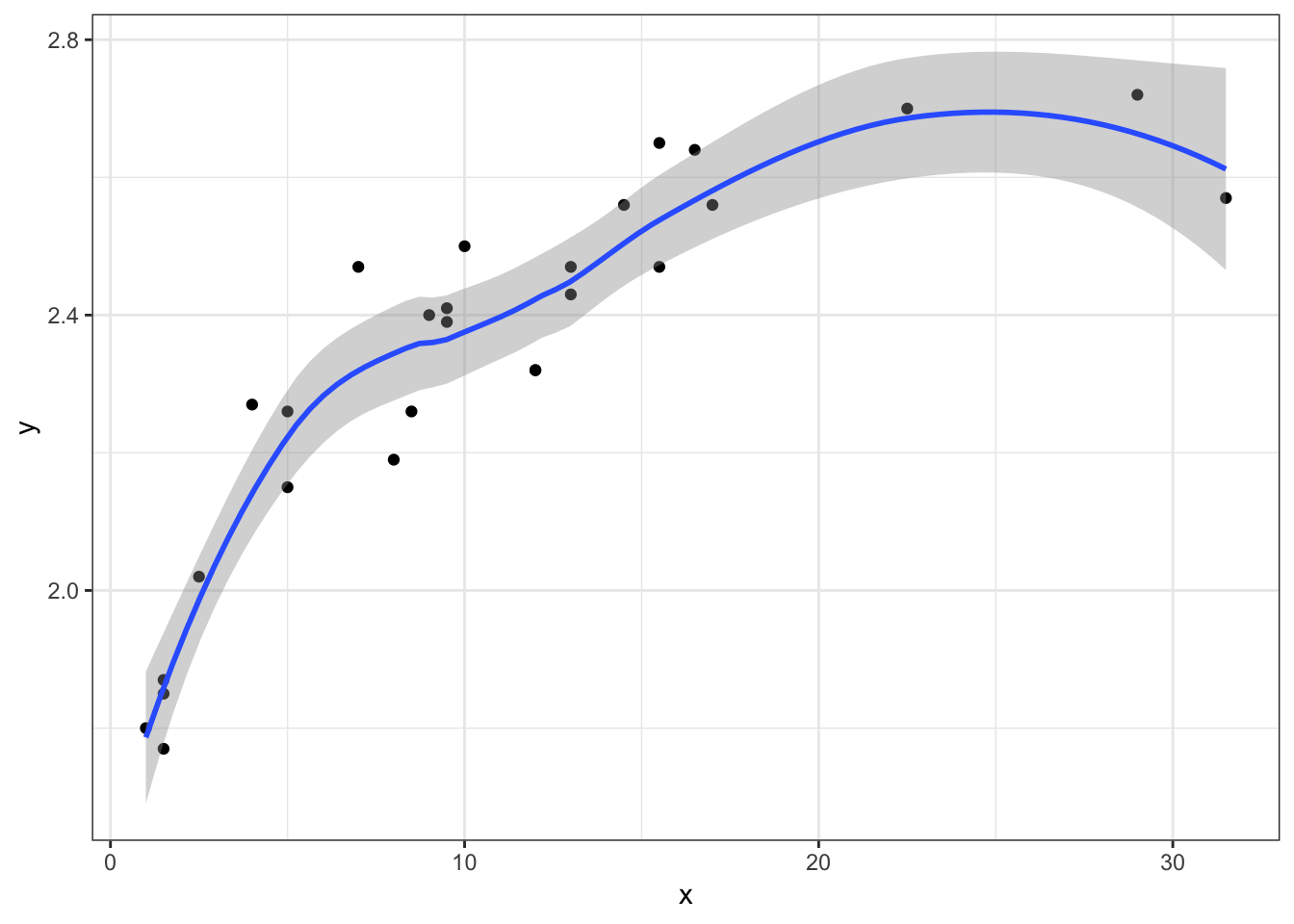
p<-ggplot(dt,aes(x,y))+geom_point()+geom_smooth()+theme_bw()
p
## `geom_smooth()` using method = 'loess' and formula 'y ~ x'
model{
for(i in 1:N){
y[i]~dnorm(mu[i],tau)
mu[i]<- alpha-beta*pow(gamma,x[i])
}
alpha~dnorm(0,0.01)
beta~dnorm(0,0.01)
gamma~ dunif(0,1)
tau ~ dgamma(0.01,0.01)
}
Implementation with R2OpenBUGS
sink("seacows.txt")
cat("
model{
for(i in 1:N){
y[i]~dnorm(mu[i],tau)
mu[i]<- alpha+beta*pow(gamma,x[i])
}
alpha~dnorm(0,0.01)
beta~dnorm(0,0.01)
gamma~ dgamma(0.01,0.01)
tau ~ dgamma(0.01,0.01)
}
",fill=T)
sink()
inits<-function(){
inits<-list(alpha=rnorm(1),beta=rnorm(1),gamma=rgamma(1,1,1),tau=rgamma(1,1,1))
}
dt<-list(x=c(seacows$x,seq(1,31.5,length=15)),
y=c(seacows$y,rep(NA,15)),
N=42)
params<-c("alpha","beta","gamma","tau","y")
filename="seacows.txt"
library(R2OpenBUGS)
outSeacows <-bugs(dt,inits,params,filename,codaPkg=F,
n.thin =3, n.iter=20000,debug=F,n.burnin = 15000,
n.chains = 2,working.directory=getwd(),
OpenBUGS.pgm=OpenBUGS.pgm, useWINE=T)
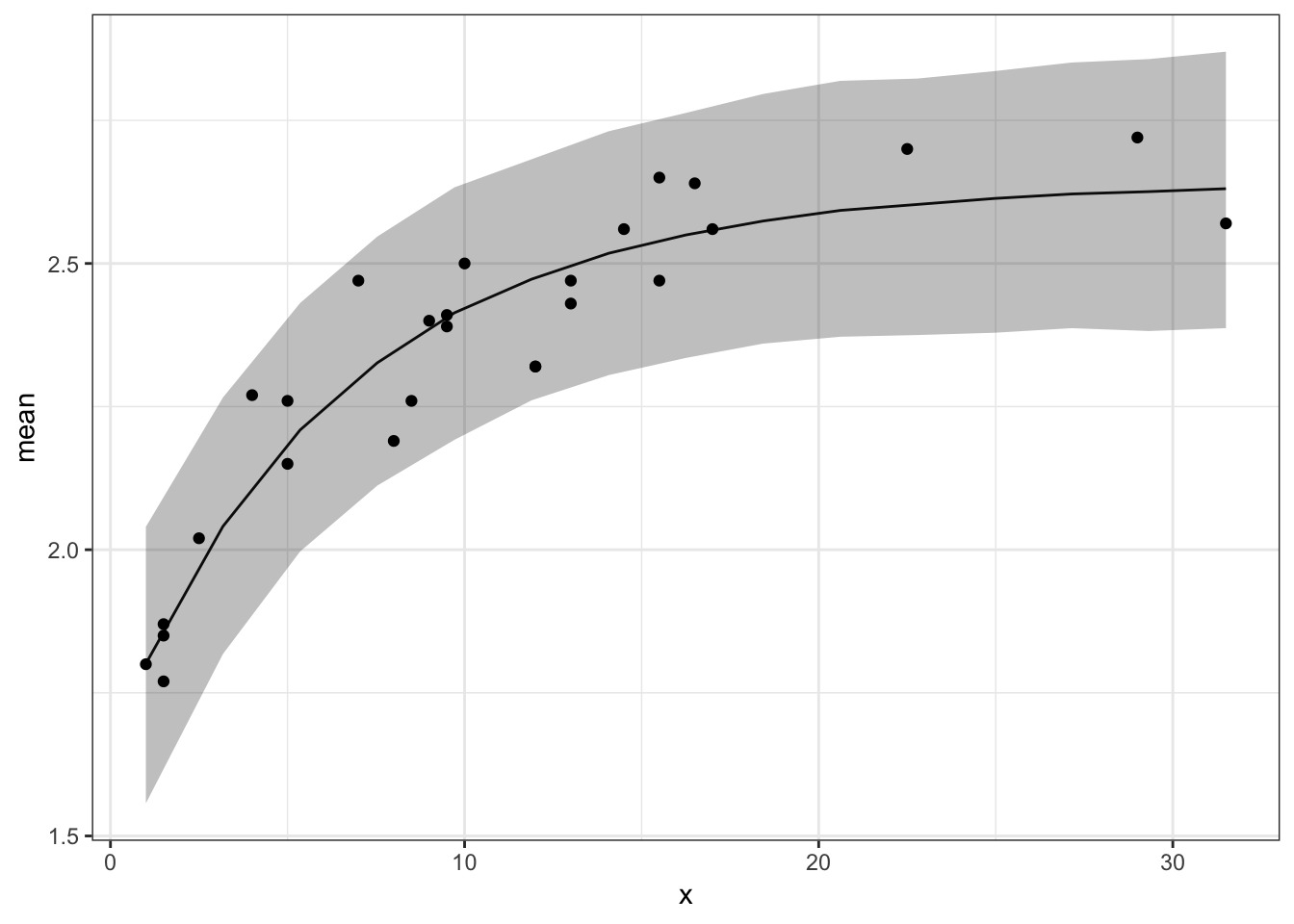
Growth Curve
dim(outSeacows$sims.array)
## [1] 5000 2 20
Y=outSeacows$summary[5:19,]
Y=Y[,c(1,3,7)]
dt_pred<-cbind(seq(1,31.5,length=15),Y)
colnames(dt_pred)=c("x","mean","Prc2.5","Prc97.5")
dt_pred=as.data.frame(dt_pred)
p<-ggplot(dt_pred,aes(x,mean))+geom_line()+geom_ribbon(aes(ymin=Prc2.5,ymax=Prc97.5),alpha=.3)
dt=t(ldply(list(seacows[[1]],seacows[[2]])))
colnames(dt)=c("x","y")
dt=as.data.frame(dt)
p<-p+geom_point(data=dt,aes(x,y))
p+theme_bw()